Wiring between close nodes in molecular networks evolves more quickly than between distant nodes
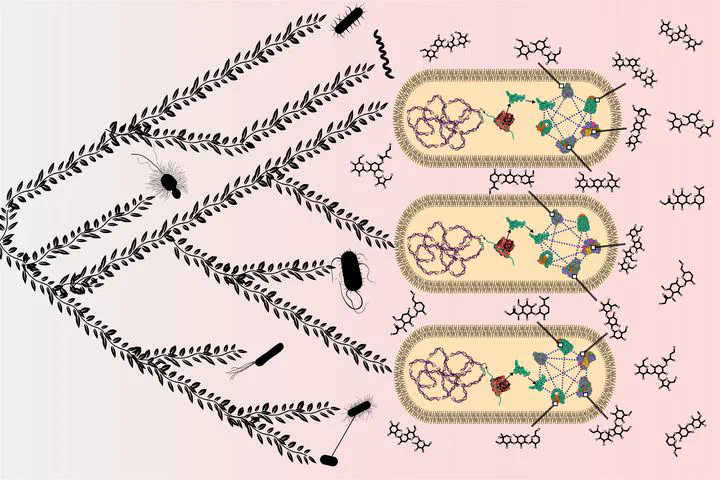 Image credit: Alejandro Gil Gomez
Image credit: Alejandro Gil GomezAbstract
As species diverge, a wide range of evolutionary processes lead to changes in protein-protein interaction networks and metabolic networks. The rate at which molecular networks evolve is an important question in evolutionary biology. Previous empirical work has focused on 18interactomes from model organisms to calculate rewiring rates, but this is limited by the relatively small number of species and sparse nature of network data across species. We present a proxy for variation in network topology: variation in drug-drug interactions (DDIs), obtained by studying drug combinations (DCs) across taxa. Here, we propose the rate at which DDIs change across species as an estimate of the rate at which the underlying molecular network changes as species diverge. We computed the evolutionary rates of DDIs using previously published data from a high throughput study in gram-negative bacteria. Using phylogenetic comparative methods, we found that DDIs diverge rapidly over short evolutionary time periods, but that divergence saturates over longer time periods. In parallel, we mapped drugs with known targets in protein-protein interaction and co-functional networks. We found that the targets of synergistic DDIs are closer in these networks than other types of DCs and that synergistic interactions have a higher evolutionary rate, meaning that nodes that are 30closer evolve at a faster rate. Future studies of network evolution may use DC data to gain 31larger-scale perspectives on the details of network evolution within and between species.
Type
Publication
Molecular Biology and Evolution